Multi-node Parallelism and Dask#
Concepts of Multi-node Jobs and Distributed Computing#
Multi-node parallelism: When a single job uses multiple nodes, it allows distributed computing across nodes to solve larger problems. This requires breaking down tasks and distributing them efficiently across nodes.
Advantages of distributed computing: Utilizing multiple nodes allows scaling workloads beyond the limits of a single machine. Examples:
Parallelizing large dataframes and arrays
Machine learning on large datasets (Training models on data that won’t fit into memory), leveraging libraries like Dask-ML
Distributed hyperparameter tuning
ETL and data pipelines
Interactive data analysis
Challenges:
Efficient distribution of tasks: Breaking tasks into smaller units that can be executed independently is crucial for maximizing parallel efficiency.
Minimizing communication overhead: Communication between nodes can slow down processing. Efficient algorithms aim to reduce the amount of inter-node communication.
Handling data dependencies: Dependencies between tasks can limit parallelism. Identifying independent tasks and minimizing dependencies are key to effective multi-node parallelism.
Introduction to Dask#
Dask collections: Dask collections are high-level abstractions that provide distributed versions of familiar data structures.
Delayed: Lazy evaluation that allows you to construct a task graph and execute it only when required. This is useful for breaking down complex computations into smaller, manageable tasks.
Dask Array: Parallelized version of NumPy arrays that can be distributed across multiple nodes.
Dask DataFrame: Parallelized version of Pandas DataFrames that can be distributed across multiple nodes.
Dask Bag: Parallelized version of Python iterators that can be distributed across multiple nodes.
Dask ML: Distributed machine learning library that can scale scikit-learn workflows to large datasets.
import dask
# Delayed: Delay computation until it is needed
def add(x, y):
return x + y
delayed_add = dask.delayed(add)(5, 6)
print(delayed_add)
Delayed('add-6c7530f5-7f69-446a-b409-24949d85484d')
result = delayed_add.compute() # Trigger computation
print(result)
11
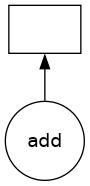
Visualizing task graphs: Dask provides visualization tools to understand the task dependencies.
# Visualize a task graph
delayed_add.visualize()
# Dask Array: Parallel NumPy arrays
import dask.array as da
import numpy as np
x = np.random.rand(10000)
y = np.random.rand(10000)
dx = da.from_array(x, chunks=len(x)//4)
dy = da.from_array(y, chunks=len(y)//4)
result = (dx + dy).sum()
print(result.compute())
9992.16562410717
# Dask DataFrame: Parallel Pandas DataFrames
import dask.dataframe as dd
import pandas as pd
df = pd.DataFrame({'x': np.random.randint(0, 10, size=10000), 'y': np.random.rand(10000)})
ddf = dd.from_pandas(df, npartitions=4)
result = ddf.groupby('x').y.mean()
print(result.compute())
x
0 0.511749
1 0.502064
2 0.492546
3 0.510551
4 0.500531
5 0.502013
6 0.497210
7 0.514784
8 0.482721
9 0.493922
Name: y, dtype: float64
# Dask Bag: Parallel Python lists
import dask.bag as db
b = db.from_sequence(range(10000), npartitions=4)
result = b.map(lambda x: x**2).filter(lambda x: x % 2 == 0).sum()
print(result.compute())
166616670000
# Create a more complex task graph for visualization
import random
def random_walk(n):
x = 0
for _ in range(n):
x += random.choice([-1, 1])
return x
b = db.from_sequence(range(1000), npartitions=4)
result = b.map(random_walk).mean()
result.visualize()
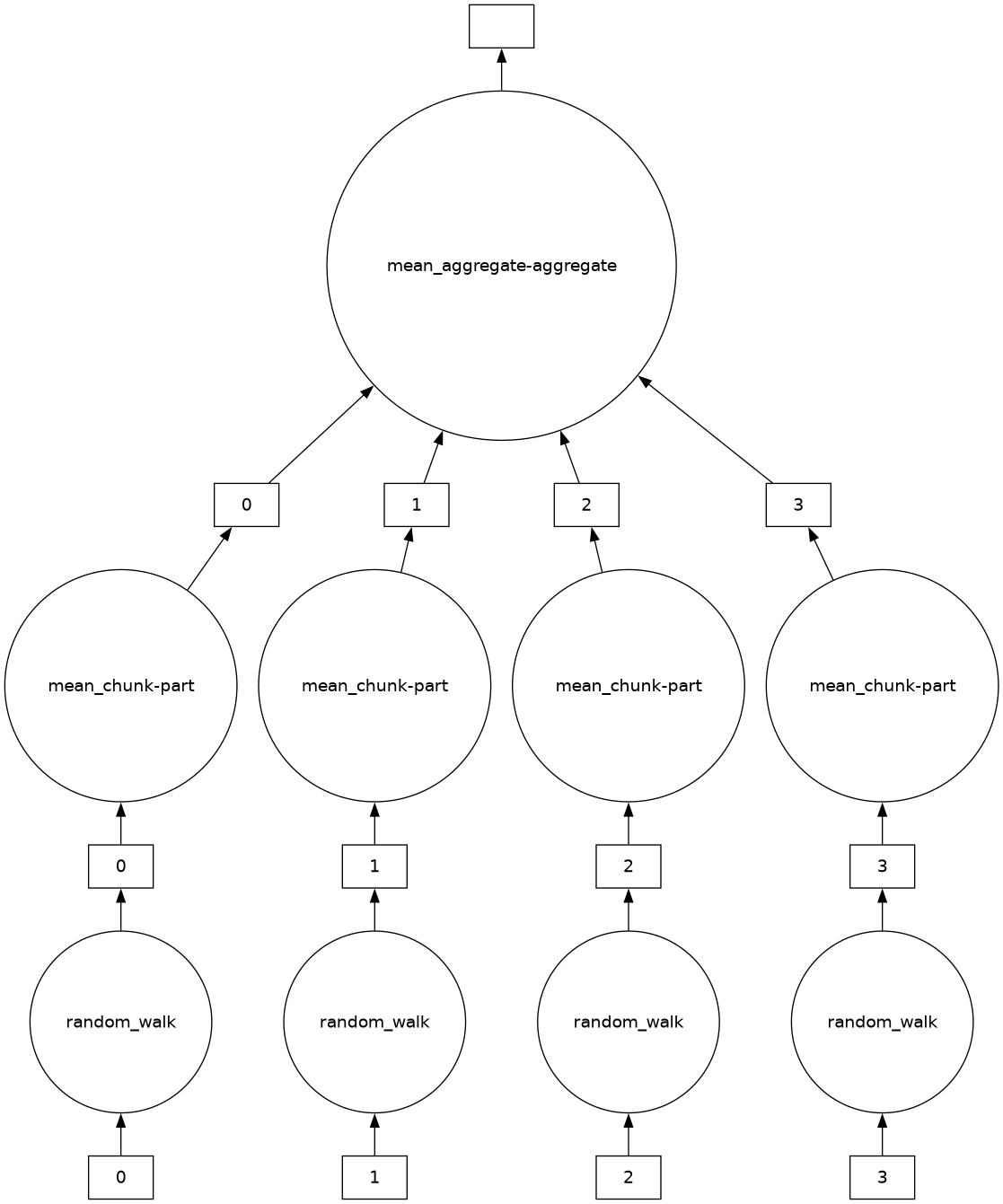
print(result.compute())
-1.5
Running Dask Jobs on Palmetto2#
Using Dask with SLURM: Dask jobs can be executed on Palmetto by leveraging SLURM to allocate nodes and manage resources. Dask can then orchestrate the distributed computation across the allocated nodes. Here’s a simple example of a SLURM script to run a Dask job:
#!/bin/bash
#SBATCH --job-name=dask_job
#SBATCH --nodes=2
#SBATCH --ntasks-per-node=2
#SBATCH --time=01:00:00
#SBATCH --output=dask_output.log
module load anaconda3 # Load Anaconda
# Ensure the conda environment is accessible across nodes
export PATH=$HOME/.conda/envs/hpc_ml/bin:$PATH
# Get the hostnames of the allocated nodes
hostnames=$(scontrol show hostnames $SLURM_JOB_NODELIST)
# Start the Dask scheduler on the first node
first_node=$(echo $hostnames | awk '{print $1}')
ssh $first_node "
module load anaconda3 &&
source activate hpc_ml &&
dask-scheduler --scheduler-file=scheduler.json &
" &
# Start Dask workers on all nodes
for node in $hostnames; do
ssh $node "
module load anaconda3 &&
source activate hpc_ml &&
dask-worker --scheduler-file=scheduler.json --nthreads=1 --nprocs=2 --memory-limit=0 &
" &
done
# Wait for all processes to complete
wait
Save the above as, e.g., dask_job.slurm, and submit it using sbatch dask_job.slurm. Then you can use ssh to connect to the first node and run your Dask code. Make sure your code points to the scheduler file (scheduler.json) to connect to the Dask cluster. For example, your code could be a .py file that looks like the following:
from dask.distributed import Client
import dask.array as da
# Connect to the Dask scheduler
client = Client(scheduler_file='scheduler.json')
# Example: Creating a random Dask array and performing a computation
x = da.random.random((10000, 10000), chunks=(1000, 1000))
result = x.mean().compute()
print("Mean of the array:", result)
If you save this as dask_example.py, you can run it by using ssh to connect to the first node running your Dask job and then running python dask_example.py.
