Regression and Classification with Fully Connected Neural Networks#
Deep learning is a large, developing field with many sub-communities, a constant stream of new developments, and unlimited application areas. Despite this complexity, most deep learning techniques share a relatively small set of algorithmic building blocks. The purpose of this notebook is to gain familiarity with some of these core concepts. Much of the intuition you develop here is applicable to the neural networks making headline news from the likes of OpenAI. To focus this notebook, we will consider the two main types of supervised machine learning:
Regression: estimation of continuous quantities
Classification: estimation of categories or discrete quantities
We will create “deep learning” models for regression and classification. We will see the power of neural networks as well as the pitfalls.
# libraries we will need
import torch
from torch import nn
import numpy as np
from torch.distributions import bernoulli
import matplotlib.pyplot as plt
# set random seeds for reproducibility
torch.manual_seed(355)
np.random.seed(355)
Tasks to solve in this notebook#
Regression#
# make some fake regression data
n_samples_reg = 100
x_reg = 1.5 * torch.randn(n_samples_reg, 1)
y_reg = 2 * x_reg + 3.0*torch.sin(3*x_reg) + 1 + 2 * torch.randn(n_samples_reg, 1)
y_reg = y_reg.squeeze()
plt.plot(x_reg, y_reg, 'o')
plt.xlabel('x')
plt.ylabel('y')
plt.grid()
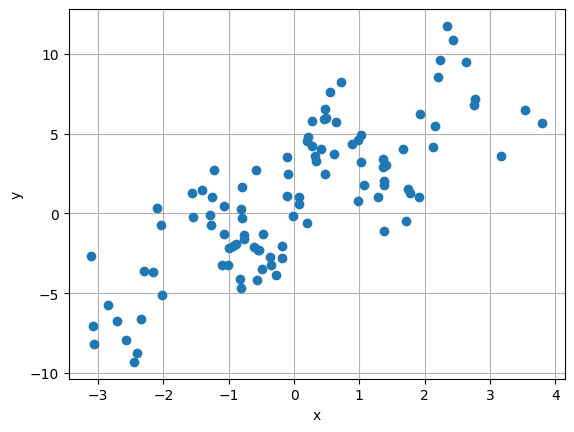
Classification#
# fake classification data
n_samples_clf = 200
x_clf = 2.5*torch.randn(n_samples_clf, 2)
d = torch.sqrt( x_clf[:, 0]**2 + x_clf[:, 1]**2 )
y_clf = d<torch.pi
# swap some labels near the boundary
width = 0.7
pgivd = 0.3 * width**2 / ((d - torch.pi)**2 + width**2)
swaps = bernoulli.Bernoulli(pgivd).sample().type(torch.bool)
y_clf[swaps] = ~y_clf[swaps]
plt.plot(x_clf[y_clf, 0], x_clf[y_clf,1], 'o', label='positive')
plt.plot(x_clf[~y_clf, 0], x_clf[~y_clf,1], 'o', label='negative')
plt.gca().set_aspect(1)
plt.grid()
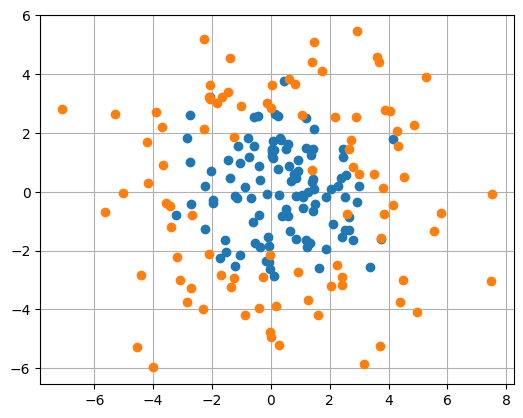
The Simplest “Deep Learning” Model: Linear Regression#
Initializing our model#
Good old fashioned linear model: \(y = wx + b\).
# Pytorch's `nn` module has lots of neural network building blocks
# nn.Linear is what we need for y=wx+b
reg_model = nn.Linear(in_features = 1, out_features = 1)
from utils import create_answer_box
create_answer_box("Why do you think ìn_features` and `out_features` are both set to 1 in the example above?", "03-01")
Why do you think ìn_featuresandout_features` are both set to 1 in the example above?
# pytorch randomly initializes the w and b
reg_model.weight, reg_model.bias
(Parameter containing:
tensor([[0.9099]], requires_grad=True),
Parameter containing:
tensor([-0.0522], requires_grad=True))
# let's look at some predictions
with torch.no_grad():
y_reg_init = reg_model(x_reg)
y_reg_init[:5]
tensor([[-0.5818],
[-1.0250],
[-0.1526],
[ 1.8780],
[ 0.4453]])
# plot the predictions before training the model
plt.plot(x_reg, y_reg, 'o', label='Actual targets')
plt.plot(x_reg, y_reg_init, 'o', label='Predicted targets')
plt.grid()
plt.legend()
_ = plt.title("This model sucks.\nWe better fit it.")
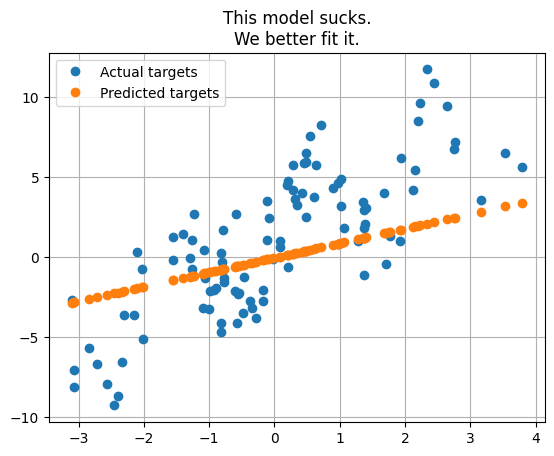
Anatomy of a training loop#
Basic idea:
Start with random parameter values
Define a loss/cost/objective measure to optimize
Use training data to evaluate the loss
Update the model to reduce the loss (use gradient descent)
Repeat until converged
Basic weight update formula: \( w_{i+1} = w_i - \mathrm{lr}\times \left.\frac{\partial L}{\partial w}\right|_{w=w_i} \)
Question
Why the minus sign in front of the second term?

# re-initialize our model each time we run this cell
# otherwise the model picks up from where it left off
reg_model = nn.Linear(in_features = 1, out_features = 1)
# the torch `optim` module helps us fit models
import torch.optim as optim
# create our optimizer object and tell it about the parameters in our model
# the optimizer will be responsible for updating these parameters during training
optimizer = optim.SGD(reg_model.parameters(), lr=0.01)
# how many times to update the model based on the available data
num_epochs = 100
# setting up a colormap for ordered colors
import matplotlib.pyplot as plt
import numpy as np
colors = plt.cm.Greens(np.linspace(0.3, 1, num_epochs // 20))
for i in range(num_epochs):
# make sure gradients are set to zero on all parameters
# by default, gradients accumulate
optimizer.zero_grad()
# "forward pass"
y_hat = reg_model(x_reg).squeeze()
# measure the loss
# this is the mean squared error
loss = torch.mean((y_hat - y_reg)**2)
# can use pytorch built-in: https://pytorch.org/docs/stable/generated/torch.nn.functional.mse_loss.html
# "backward pass"
# that is, compute gradient of loss wrt all parameters
loss.backward()
# parameter updates
# use the basic weight update formula given above (with slight modifications)
optimizer.step()
if i % 20 == 0:
print(f"Epoch {i+1}. MSE Loss = {loss:0.3f}")
with torch.no_grad():
y_reg_init = reg_model(x_reg)
ix = x_reg.argsort(axis=0).squeeze()
plt.plot(x_reg.squeeze()[ix], y_reg_init.squeeze()[ix], '-', color=colors[i // 20], label=i, alpha=1)
plt.plot(x_reg, y_reg, 'ok', label="Actual data")
plt.grid()
_ = plt.legend(title='Epoch')
Epoch 1. MSE Loss = 22.410
Epoch 21. MSE Loss = 9.624
Epoch 41. MSE Loss = 7.960
Epoch 61. MSE Loss = 7.742
Epoch 81. MSE Loss = 7.713
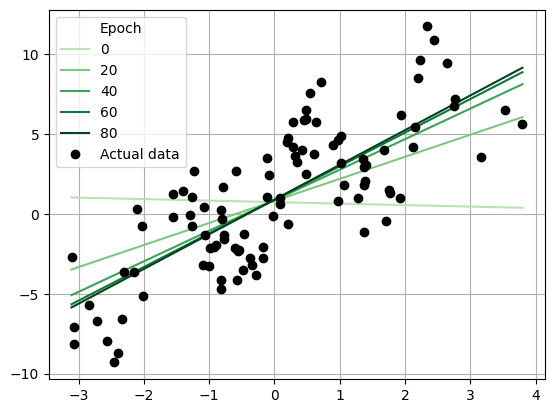
import torch
import torch.nn as nn
import torch.optim as optim
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.animation import FuncAnimation
from IPython.display import HTML
# re-initialize our model each time we run this cell
# otherwise the model picks up from where it left off
reg_model = nn.Linear(in_features=1, out_features=1)
# create our optimizer object and tell it about the parameters in our model
# the optimizer will be responsible for updating these parameters during training
optimizer = optim.SGD(reg_model.parameters(), lr=0.01)
# how many times to update the model based on the available data
num_epochs = 100
# setup the figure for animation
fig, ax = plt.subplots()
# plot the initial data points
ax.plot(x_reg, y_reg, 'ok', label="Actual data")
line, = ax.plot([], [], 'g-', label="Regression Line")
epoch_text = ax.text(0.02, 0.95, '', transform=ax.transAxes)
plt.grid()
plt.legend()
# function to initialize the animation
def init():
line.set_data([], [])
epoch_text.set_text('')
return line, epoch_text
# function to update the plot during each frame of the animation
def update(epoch):
# make sure gradients are set to zero on all parameters
# by default, gradients accumulate
optimizer.zero_grad()
# "forward pass"
y_hat = reg_model(x_reg).squeeze()
# measure the loss
# this is the mean squared error
loss = torch.mean((y_hat - y_reg)**2)
# "backward pass"
# that is, compute gradient of loss wrt all parameters
loss.backward()
# parameter updates
optimizer.step()
if epoch % 20 == 0:
print(f"Epoch {epoch+1}. MSE Loss = {loss:0.3f}")
# update the regression line
with torch.no_grad():
y_reg_init = reg_model(x_reg)
ix = x_reg.argsort(axis=0).squeeze()
line.set_data(x_reg.squeeze()[ix], y_reg_init.squeeze()[ix])
# update the epoch text
epoch_text.set_text(f'Epoch: {epoch+1}')
return line, epoch_text
# create the animation
ani = FuncAnimation(fig, update, frames=num_epochs, init_func=init, blit=True)
# display the animation in Jupyter
plt.close(fig) # prevent static image from displaying
HTML(ani.to_jshtml())
Epoch 1. MSE Loss = 20.623
Epoch 21. MSE Loss = 10.099
Epoch 41. MSE Loss = 8.337
Epoch 61. MSE Loss = 7.931
Epoch 81. MSE Loss = 7.800
Question
What shortcomings does this model have?
🔥 IMPORTANT CONCEPT 🔥
Model bias is a type of error that occurs when your model doesn't have enough flexibility to represent the real-world data!
Linear classification#
A slight modification to the model#
We need to modify our model because the outputs are true/false
y_clf.unique()
tensor([False, True])
While training the model, we cannot simply threshold the model outputs (e.g. call outputs greater than 0 ‘True’). Instead, we can have our model output a continuous number between 0 and 1. Use sigmoid to transform the output of a linear model to the range (0,1): $\( p(y=1 | \vec{x}) = \mathrm{sigmoid}(\vec w \cdot \vec x + b) \)$
Question
Why doesn't thresholding work while training?
🔥 IMPORTANT CONCEPT 🔥
In order to use gradient descent, neural networks must be differentiable.
# what is sigmoid?
x_plt = torch.linspace(-10,10, 100)
y_plt = torch.sigmoid(x_plt)
plt.axvline(0, color='gray')
plt.plot(x_plt, y_plt)
plt.grid()
plt.xlabel('input')
plt.ylabel('output')
plt.gcf().set_size_inches(5,2.5)
_ = plt.title('Sigmoid Function')
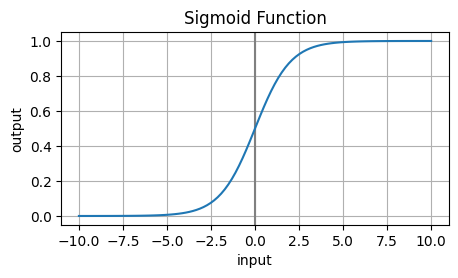
The sigmoid function is defined as:
# proposed new model for classification
# use nn.Sequential to string operations together
model_clf = nn.Sequential(
nn.Linear(2, 1), # notice: we now have two inputs
nn.Sigmoid() # sigmoid squishes real numbers into (0, 1)
)
# what do the outputs look like?
with torch.no_grad():
print(model_clf(x_clf)[:10])
# notice how they all lie between 0 and 1
# we loosely interpret these numbers as
# "the probability that y=1 given the provided value of x"
tensor([[0.5043],
[0.2114],
[0.4564],
[0.2263],
[0.2401],
[0.2266],
[0.7676],
[0.4448],
[0.6703],
[0.2872]])
# turning probabilities into "decisions"
# apply a threshold
with torch.no_grad():
print(model_clf(x_clf)[:10] > 0.5)
tensor([[ True],
[False],
[False],
[False],
[False],
[False],
[ True],
[False],
[ True],
[False]])
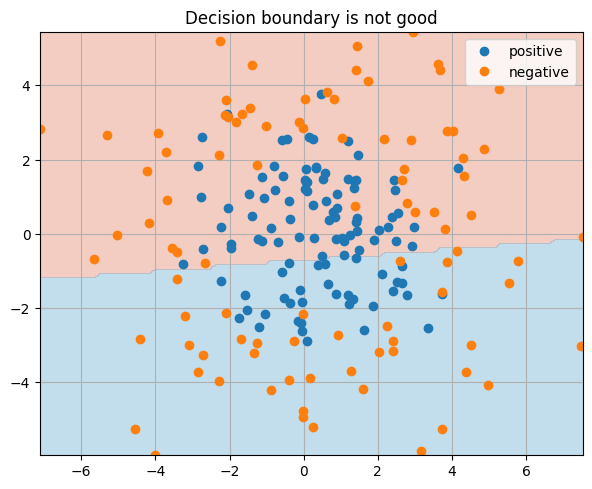
# let's see how this model does before any training
# Code to plot the decision regions
xx, yy = mg = torch.meshgrid(
torch.linspace(x_clf[:,0].min(), x_clf[:,0].max(), 100),
torch.linspace(x_clf[:,1].min(), x_clf[:,1].max(), 100)
)
grid_pts = torch.vstack([xx.ravel(), yy.ravel()]).T
with torch.no_grad():
# use a 0.5 decision threshold
grid_preds = model_clf(grid_pts).squeeze()>0.5
grid_preds = grid_preds.reshape(xx.shape)
plt.gcf().set_size_inches(7,7)
plt.plot(x_clf[y_clf, 0], x_clf[y_clf,1], 'o', label='positive')
plt.plot(x_clf[~y_clf, 0], x_clf[~y_clf,1], 'o', label='negative')
plt.gca().set_aspect(1)
plt.grid()
plt.contourf(xx, yy, grid_preds, cmap=plt.cm.RdBu, alpha=0.4)
plt.legend()
_ = plt.title("Decision boundary is not good")
/home/cehrett/.conda/envs/PytorchWorkshop/lib/python3.11/site-packages/torch/functional.py:539: UserWarning: torch.meshgrid: in an upcoming release, it will be required to pass the indexing argument. (Triggered internally at /pytorch/aten/src/ATen/native/TensorShape.cpp:3637.)
return _VF.meshgrid(tensors, **kwargs) # type: ignore[attr-defined]
Training loop#
The main difference between regression and classification is the loss function. For regression, we used mean squared error. For classification, we will use cross-entropy loss. This loss formula encourages the model to output low values for \(p(y=1|x)\) when the class is 0 and high values when the class is 1.
# re-initialize our model each time we run this cell
# otherwise the model picks up from where it left off
model_clf = nn.Sequential(
nn.Linear(2, 1),
nn.Sigmoid()
)
# create our optimizer object and tell it about the parameters in our model
optimizer = optim.SGD(model_clf.parameters(), lr=0.01)
# torch needs the class labels to be zeros and ones, not False/True
y_clf_int = y_clf.type(torch.int16)
# how many times to update the model based on the available data
num_epochs = 1000
for i in range(num_epochs):
optimizer.zero_grad()
y_hat = model_clf(x_clf).squeeze()
# measure the goodness of fit
# need to use a different loss function here
# this is the "cross entropy loss"
loss = -y_clf_int * torch.log(y_hat) - (1-y_clf_int)*torch.log(1-y_hat)
loss = loss.mean()
# pytorch built-in: https://pytorch.org/docs/stable/generated/torch.nn.functional.binary_cross_entropy.html
# update the model
loss.backward() # gradient computation
optimizer.step() # weight updates
if i % 100 == 0:
print(f"Epoch {i+1}. CE Loss = {loss:0.3f}")
Epoch 1. CE Loss = 0.845
Epoch 101. CE Loss = 0.701
Epoch 201. CE Loss = 0.691
Epoch 301. CE Loss = 0.690
Epoch 401. CE Loss = 0.690
Epoch 501. CE Loss = 0.690
Epoch 601. CE Loss = 0.690
Epoch 701. CE Loss = 0.690
Epoch 801. CE Loss = 0.690
Epoch 901. CE Loss = 0.690
with torch.no_grad():
# use a 0.5 decision threshold
grid_preds = model_clf(grid_pts).squeeze()>0.5
grid_preds = grid_preds.reshape(xx.shape)
plt.gcf().set_size_inches(7,7)
plt.plot(x_clf[y_clf, 0], x_clf[y_clf,1], 'o', label='positive')
plt.plot(x_clf[~y_clf, 0], x_clf[~y_clf,1], 'o', label='negative')
plt.gca().set_aspect(1)
plt.grid()
plt.contourf(xx, yy, grid_preds, cmap=plt.cm.RdBu, alpha=0.4)
plt.legend()
_ = plt.title("Decision boundary is still not good")
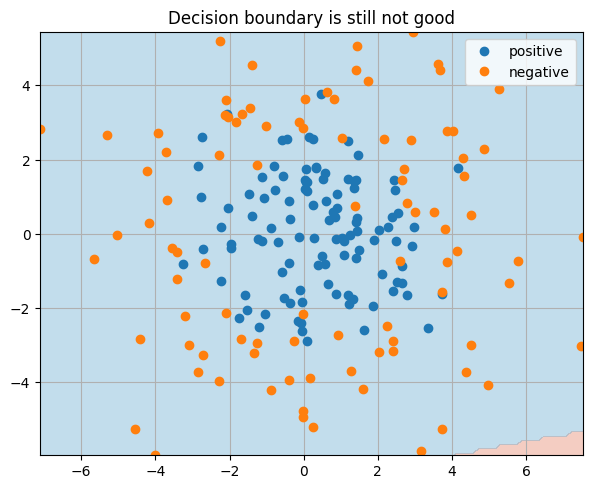
Question
Why did this experiment fail? Any ideas what we could do to make it work?
Summing up linear models#
Sometimes linear models can be a good approximation to our data. Sometimes not.
Linear models tend to be biased in the statistical sense of the word. That is, they enforce linearity in the form of linear regression lines or linear decision boundaries. The linear model will fail to the degree that the real-world data is non-linear.
Getting non-linear with fully-connected neural networks#
We can compose simple math operations together to represent very complex functions.

A few helper functions#
def train_and_test_regression_model(model, num_epochs=1000, reporting_interval=100):
# Split data into train/test sets (80/20 split)
n_samples = len(x_reg)
torch.manual_seed(355) # for reproducibility
indices = torch.randperm(n_samples)
train_size = int(0.8 * n_samples)
train_indices = indices[:train_size]
test_indices = indices[train_size:]
x_train, y_train = x_reg[train_indices], y_reg[train_indices]
x_test, y_test = x_reg[test_indices], y_reg[test_indices]
# create our optimizer object and tell it about the parameters in our model
optimizer = optim.SGD(model.parameters(), lr=0.01)
# how many times to update the model based on the available data
for i in range(num_epochs):
optimizer.zero_grad()
y_hat = model(x_train).squeeze()
# measure the goodness of fit on training data
loss = torch.mean((y_hat - y_train)**2)
# update the model
loss.backward() # gradient computation
optimizer.step() # weight updates
if i % reporting_interval == 0:
print(f"Epoch {i+1}. Training MSE Loss = {loss:0.3f}")
# Evaluate on test set
with torch.no_grad():
y_train_pred = model(x_train)
y_test_pred = model(x_test)
test_mse = torch.mean((y_test_pred.squeeze() - y_test)**2)
print(f"Final Test MSE: {test_mse:0.3f}")
plt.plot(x_train.cpu(), y_train.cpu(), 'o', label='Training data')
plt.plot(x_test.cpu(), y_test.cpu(), 's', label='Test data')
plt.plot(x_train.cpu(), y_train_pred.cpu(), 'x', label='Training predictions')
plt.plot(x_test.cpu(), y_test_pred.cpu(), '+', label='Test predictions')
plt.grid()
plt.legend()
return test_mse.item()
def train_and_test_classification_model(model, num_epochs=1000, reporting_interval=100):
# Split data into train/test sets (80/20 split)
n_samples = len(x_clf)
torch.manual_seed(355) # for reproducibility
indices = torch.randperm(n_samples)
train_size = int(0.8 * n_samples)
train_indices = indices[:train_size]
test_indices = indices[train_size:]
x_train, y_train = x_clf[train_indices], y_clf_int[train_indices]
x_test, y_test = x_clf[test_indices], y_clf_int[test_indices]
# create our optimizer object and tell it about the parameters in our model
optimizer = optim.SGD(model.parameters(), lr=0.01)
# how many times to update the model based on the available data
for i in range(num_epochs):
optimizer.zero_grad()
y_hat = model(x_train).squeeze()
# measure the goodness of fit on training data
loss = -y_train * torch.log(y_hat) - (1-y_train)*torch.log(1-y_hat)
loss = loss.mean()
# update the model
loss.backward() # gradient computation
optimizer.step() # weight updates
if i % reporting_interval == 0:
print(f"Epoch {i+1}. Training CE Loss = {loss:0.3f}")
# Evaluate on test set
with torch.no_grad():
y_test_pred = model(x_test).squeeze()
test_predictions = (y_test_pred > 0.5).int()
test_accuracy = (test_predictions == y_test).float().mean()
print(f"Final Test Accuracy: {test_accuracy:0.3f}")
grid_preds = model(grid_pts).squeeze()>0.5
grid_preds = grid_preds.reshape(xx.shape)
plt.gcf().set_size_inches(7,7)
plt.plot(x_clf[y_clf, 0], x_clf[y_clf,1], 'o', label='positive')
plt.plot(x_clf[~y_clf, 0], x_clf[~y_clf,1], 'o', label='negative')
plt.gca().set_aspect(1)
plt.grid()
plt.contourf(xx, yy, grid_preds, cmap=plt.cm.RdBu, alpha=0.4)
plt.legend()
return test_accuracy.item()
Regression#
What about nested linear models? Might this do something interesting? $\( y = w_1(w_0 x + b_0) + b_1 \)$
# We can build more complex models by stringing together more operations inside nn.Sequential:
model = nn.Sequential(
nn.Linear(in_features = 1, out_features = 1),
nn.Linear(1,1)
)
train_and_test_regression_model(model)
_ = plt.title("But wait, that's still just a line?!")
Epoch 1. Training MSE Loss = 19.467
Epoch 101. Training MSE Loss = 7.961
Epoch 201. Training MSE Loss = 7.961
Epoch 301. Training MSE Loss = 7.961
Epoch 401. Training MSE Loss = 7.961
Epoch 501. Training MSE Loss = 7.961
Epoch 601. Training MSE Loss = 7.961
Epoch 701. Training MSE Loss = 7.961
Epoch 801. Training MSE Loss = 7.961
Epoch 901. Training MSE Loss = 7.961
Final Test MSE: 6.824
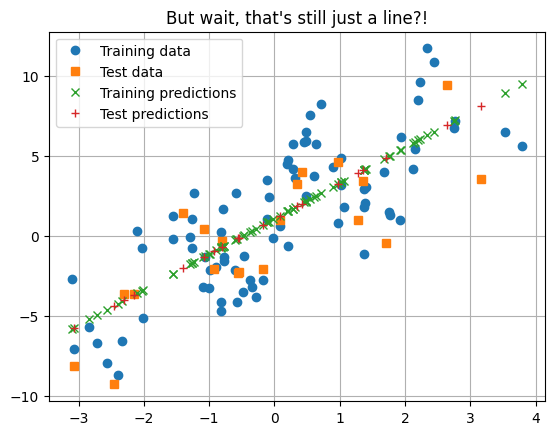
Question
Why is it still just a line? How can we fix it?
# We need a non-linearity between the two linear operations
model = nn.Sequential(
nn.Linear(in_features = 1, out_features = 1),
nn.Tanh(),
nn.Linear(1,1)
)
# this model does not reduce to a linear operation
train_and_test_regression_model(model)
_ = plt.title("Non-linearity! But not enough.")
Epoch 1. Training MSE Loss = 23.139
Epoch 101. Training MSE Loss = 10.007
Epoch 201. Training MSE Loss = 8.227
Epoch 301. Training MSE Loss = 8.137
Epoch 401. Training MSE Loss = 8.119
Epoch 501. Training MSE Loss = 8.106
Epoch 601. Training MSE Loss = 8.096
Epoch 701. Training MSE Loss = 8.087
Epoch 801. Training MSE Loss = 8.080
Epoch 901. Training MSE Loss = 8.074
Final Test MSE: 9.127
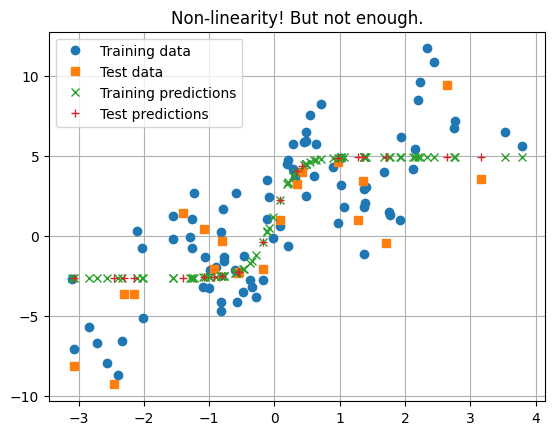
# Let's really crank up the number of hidden nodes
model = nn.Sequential(
nn.Linear(in_features = 1, out_features = 30),
nn.Tanh(),
nn.Linear(30,30),
nn.Tanh(),
nn.Linear(30,30),
nn.Tanh(),
nn.Linear(30,30),
nn.Tanh(),
nn.Linear(30,30),
nn.Tanh(),
nn.Linear(30,30),
nn.Tanh(),
nn.Linear(30,30),
nn.Tanh(),
nn.Linear(30,1)
)
train_and_test_regression_model(model, num_epochs=20_000, reporting_interval=2_000)
Epoch 1. Training MSE Loss = 22.043
Epoch 2001. Training MSE Loss = 3.423
Epoch 4001. Training MSE Loss = 2.753
Epoch 6001. Training MSE Loss = 2.431
Epoch 8001. Training MSE Loss = 2.325
Epoch 10001. Training MSE Loss = 2.185
Epoch 12001. Training MSE Loss = 2.090
Epoch 14001. Training MSE Loss = 2.450
Epoch 16001. Training MSE Loss = 1.883
Epoch 18001. Training MSE Loss = 2.208
Final Test MSE: 3.188
3.1882119178771973
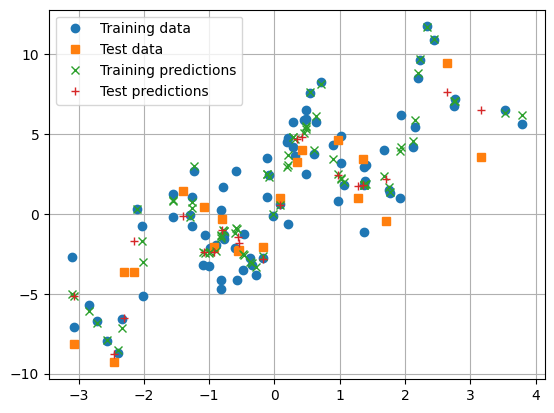
Question
What is wrong with this picture?
with torch.no_grad():
x_grid = torch.linspace(x_reg.min(), x_reg.max(), 200)[:,None]
y_reg_grid = model(x_grid)
y_reg_init = model(x_reg)
plt.plot(x_reg.cpu(), y_reg.cpu(), 'o', label='Actual targets')
plt.plot(x_reg.cpu(), y_reg_init.cpu(), 'x', label='Predicted targets')
plt.plot(x_grid.cpu(), y_reg_grid.cpu(), 'k-', label='Predicted')
plt.grid()
plt.legend()
<matplotlib.legend.Legend at 0x7fa9986fd3d0>
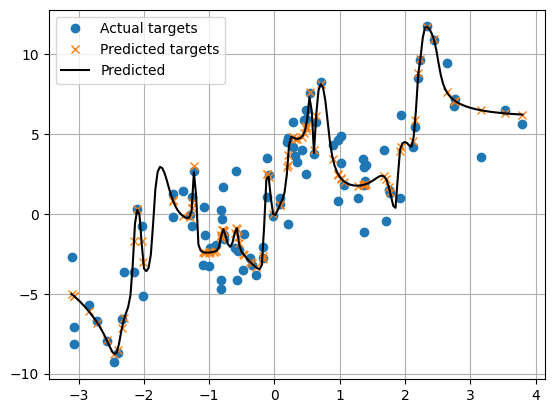
Question
Where is overfitting most severe? How might having more data alleviate the problem?
🔥 IMPORTANT CONCEPT 🔥
Overfitting is a type of error that occurs when your model memorizes the training samples and fails to generalize to unseen data. This usually ocurrs when the model has excess capacity.
😅 EXERCISE 😅
Using the code cell below, design and test a model that gets it "just right".
# model = your model goes here
# train_and_test_regression_model(model, num_epochs=10000, reporting_interval=1000)
from utils import create_answer_box
create_answer_box("What is the final test MSE of your model?", "03-02")
create_answer_box("Please copy/paste your model definition code here.", "03-03")
Classification#
Let’s apply the same non-linear treatment to the case of classification.
model = nn.Sequential(
nn.Linear(in_features = 2, out_features = 2),
nn.Tanh(),
nn.Linear(2,1),
nn.Sigmoid()
)
train_and_test_classification_model(model, 10_000, 1000)
_ = plt.title("Not non-linear enough")
Epoch 1. Training CE Loss = 0.750
Epoch 1001. Training CE Loss = 0.686
Epoch 2001. Training CE Loss = 0.679
Epoch 3001. Training CE Loss = 0.674
Epoch 4001. Training CE Loss = 0.666
Epoch 5001. Training CE Loss = 0.646
Epoch 6001. Training CE Loss = 0.615
Epoch 7001. Training CE Loss = 0.579
Epoch 8001. Training CE Loss = 0.550
Epoch 9001. Training CE Loss = 0.530
Final Test Accuracy: 0.725
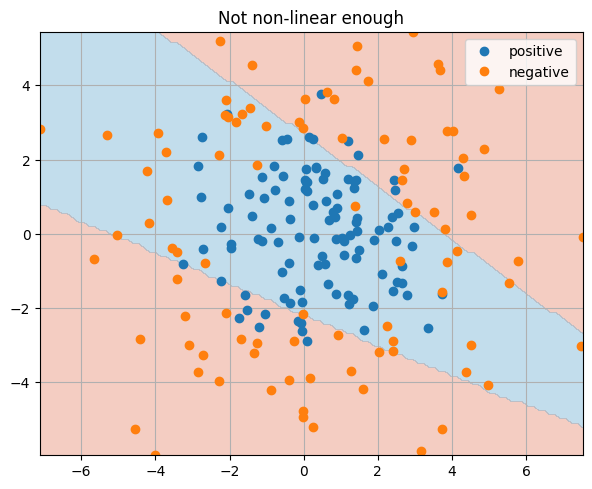
Question
What would overfitting look like in this type of graph?
model = nn.Sequential(
nn.Linear(in_features = 2, out_features = 50),
nn.Tanh(),
nn.Linear(50,50),
nn.Tanh(),
nn.Linear(50,50),
nn.Tanh(),
nn.Linear(50,50),
nn.Tanh(),
nn.Linear(50,50),
nn.Tanh(),
nn.Linear(50,50),
nn.Tanh(),
nn.Linear(50,1),
nn.Sigmoid()
)
train_and_test_classification_model(model, 30000, 3000)
_ = plt.title("What went wrong here?")
Epoch 1. Training CE Loss = 0.690
Epoch 3001. Training CE Loss = 0.243
Epoch 6001. Training CE Loss = 0.216
Epoch 9001. Training CE Loss = 0.203
Epoch 12001. Training CE Loss = 0.186
Epoch 15001. Training CE Loss = 0.169
Epoch 18001. Training CE Loss = 0.155
Epoch 21001. Training CE Loss = 0.140
Epoch 24001. Training CE Loss = 0.120
Epoch 27001. Training CE Loss = 0.100
Final Test Accuracy: 0.800
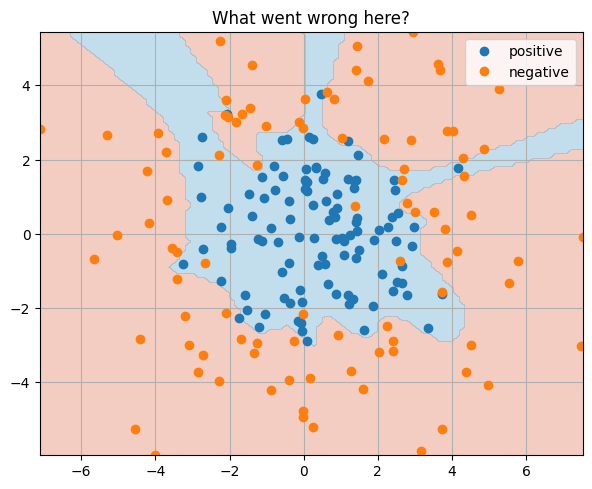
Question
What's wrong with this picture?
😅 EXERCISE 😅
Using the code cell below, design and test a model that gets it "just right".
# model = your model goes here
# train_and_test_classification_model(model, 20000, 2000)
from utils import create_answer_box
create_answer_box("What is the final test accuracy?", "03-04")
create_answer_box("Please copy/paste your model definition code here.", "03-05")
Summing up fully connected neural networks#
Fully connected neural networks can represent highly non-linear functions
We can learn good functions through gradient descent
Overfitting is a big problem
These concepts apply to nearly any neural network trained with gradient descent from the smallest fully connected net to GPT-4.
